News
Members
Publications
Software / Data
Job offers
Images / Videos
Collaborations
Conferences
Lab meetings: "Les partages de midi"
Practical information
Members Area
Next conferences we are in …



This projet is in the continuity of Ammar Mechouche's PhD on the semantic annotation of cortical structures. The general objective remains to produce image annotations that enable to associate explicit semantics to anatomical features extracted from anatomical MRI images and Diffusion MRI images. Such features can be regions of interest (ROI) representing anatomical structures such as gyri or gyri parts, as well as fiber bundles extracted from Diffusion MRI images, using deterministic or probabilistic tractography methods. The annotations refer to ontologies, that provide a reference terminology and represent the common properties of the referred entities. In our vision, such an approach can be used for various purposes: (1) reasoning - e.g. spatial reasoning - about anatomical entities, especially in the context of surgery planning; (2) intelligent querying of image repositories, and (3) mediation for the integration of heterogeneous databases.
connectomics, ontology, connectome, tractography, data sharing, neuroimaging, neuroanatomy, semantic web
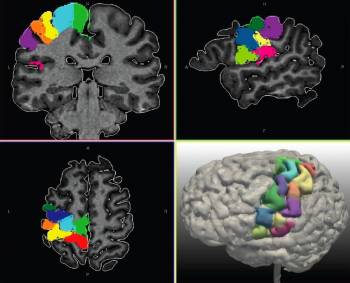
In a preliminary phase of the project (Master student work of Elsa Magro in 2010) we investigated in a population of 20 healthy subjects the connections within the central area, based on a ROI parcellation deduced from the sulcal folds (figure 1).
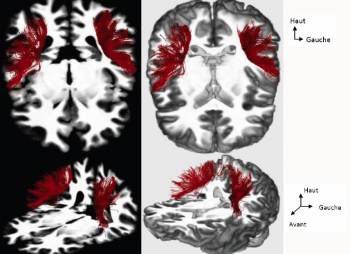
This study brought interesting results about U-fibers connecting the precentral and postcentral gyri (figure 2).
In the following of the project we propose an ontology called “Human Connectomics Ontology” (HCO) and we show how it can be used to annotate in vivo human connectomics datasets. This ontology is an extension of the Foundational Model of Anatomy (FMA), specifically designed to represent structural connectivity imaged by diffusion tractography. It includes a subset of FMA, called Neuro-DL-FMA, built from several lists of brain anatomical structures: (1) the Freesurfer cortical regions (Desikan et al. 2006), (2) the white matter bundles of the JHU white matter tractography atlas (Wakana et al. 2007) and (3) a list of subcortical and hippocampal structures. In Neuro-DL-FMA, the original object properties (such as fma:regional_part_of and fma:constitutional_part_of) - originally represented at the individuals' level - were translated at the classes' level (using existential restriction axioms). The HCO ontology introduces several classes and object properties to denote entities that participate in structural connections reconstructed by diffusion tractography, namely:
Experiments have been conducted to show how HCO could assist in analyzing real connectomics patterns. The following annotation files describe connectomics datasets describing connections in the right medial Brodmann Area 6 in five healthy subjects.